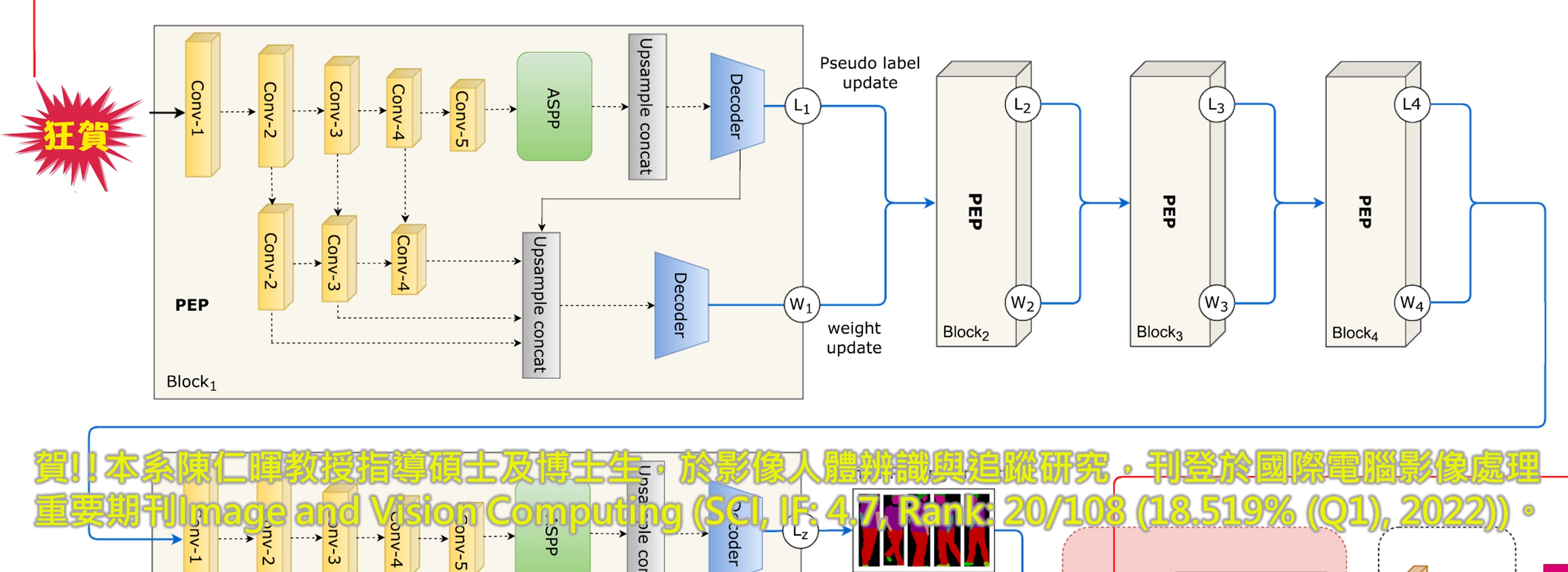
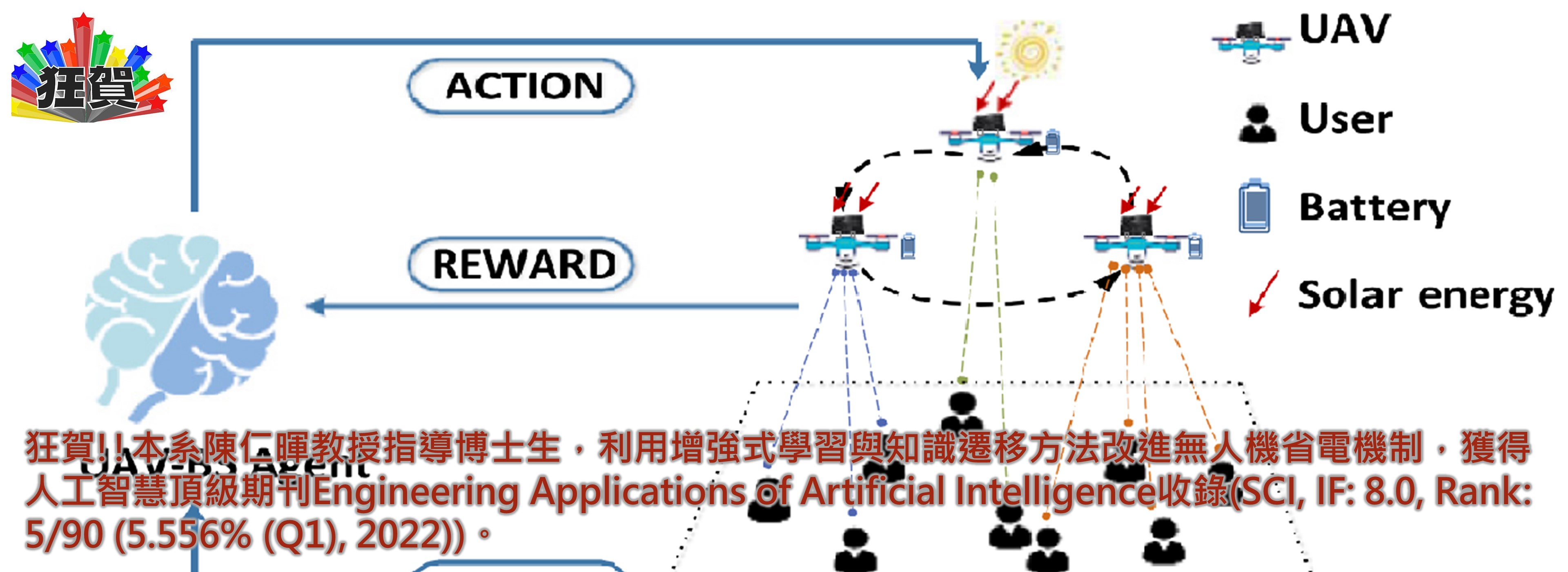
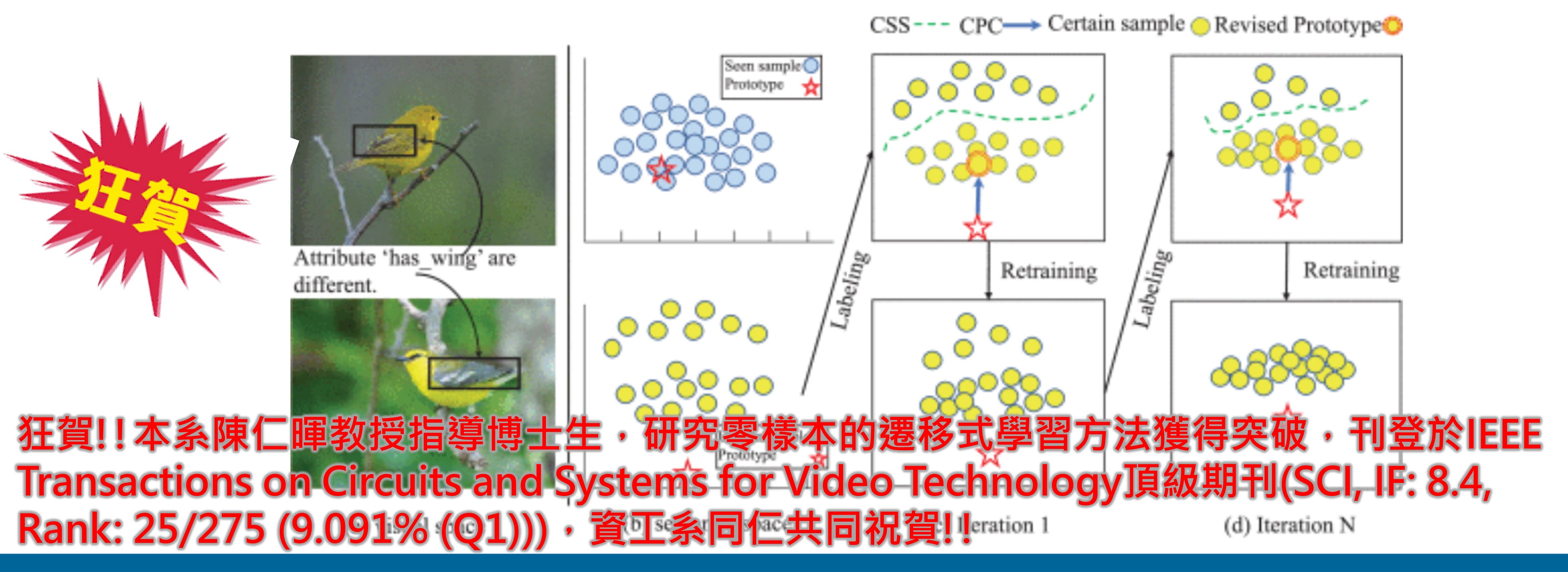
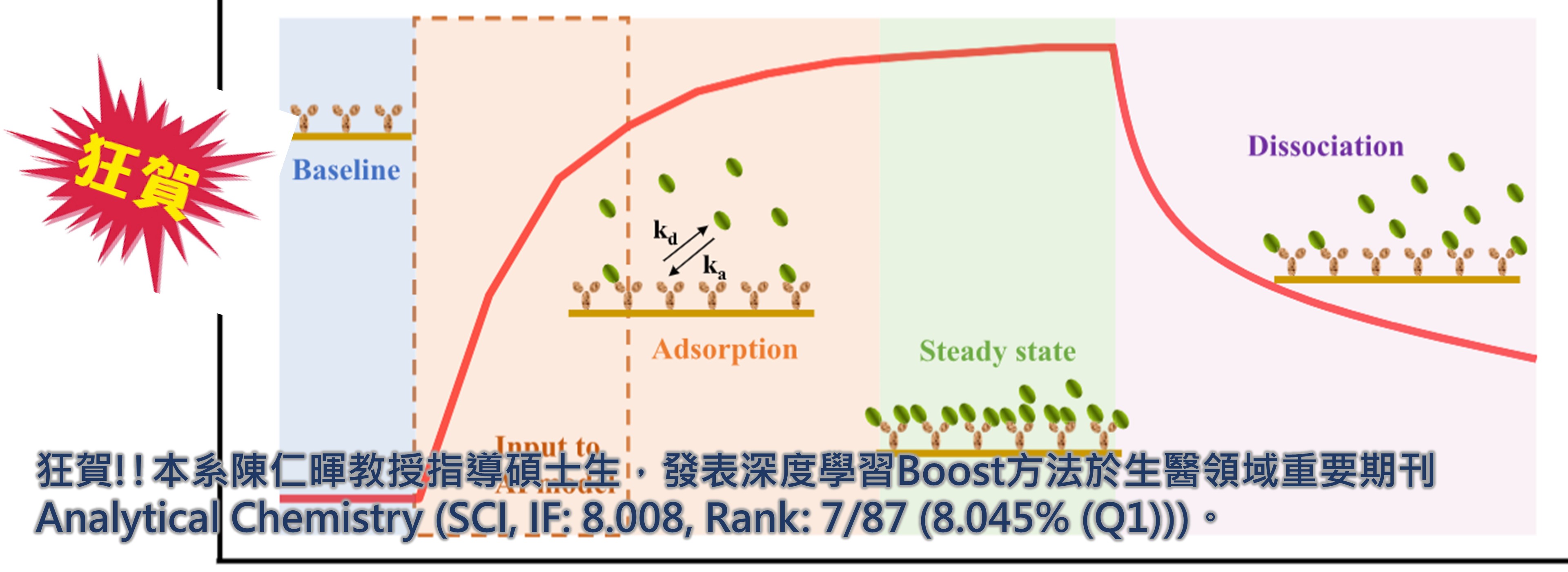
狂賀!!本系陳仁暉教授指導碩士生,發表深度學習Boost方法於生醫領域重要期刊Analytical Chemistry 。
►狂賀!!本系陳仁暉教授指導碩士生,發表深度學習Boost方法於生醫領域重要期刊Analytical Chemistry (SCI, IF: 8.008, Rank: 7/87 (8.045% (Q1)))。
Ying-Feng Chang, Sin-You Chen, Chi-Ching Lee, Jenhui Chen*, and Chao-Sung Lai*, "Easy and Rapid Approach to Obtaining the Binding Affinity of Biomolecular Interactions based on the Deep Learning Boost," Analytical Chemistry, vol. 29, no. 24, pp. 10427-10434, July 2022. (SCI, IF: 8.008, Rank: 7/87 (8.045% (Q1), 2021) in Analytical Chemistry) DOI: 10.1021/acs.analchem.2c01620
Recently, the deep learning (DL) dimension of artificial intelligence has received much attention from biochemical researchers and thus has gradually become the key approach adopted in the area of biosensing applications. Studies have shown that the use of DL techniques for sensing can not only shorten the time of data analysis but also significantly increase the accuracy of data analysis and prediction, resulting in the performance improvement of biosensing systems in comparison to conventional methods. However, obtaining reliable equilibrium and rate constants of biomolecular interactions during the detection process remains difficult and time-consuming to date. In this study, we propose a transformed model based on the deep transfer learning and sequence-to-sequence autoencoder that can successfully transfer the SPR sensorgram to the protein-binding constants, that is, the association rate constant (ka) and dissociation rate constant (kd), which provide crucial information to understand the mechanisms of drug action and the functional structures of biomolecules. Experimentally, we first trained and tested the pre-trained model using the Langmuir model which generated ideal SPR sensorgrams and then we fine-tuned the pre-trained model through the augmented SPR sensorgrams which were synthesized by using the synthesized minority oversampling technique (SMOTE) through the moderate-scale experiment. Next, the fine-tuned model was inputted with a short experimental SPR sensorgram that only needs 110 s, and the sensorgram was directly transformed into a reconstructed ideal sensorgram. Finally, the binding kinetic constants, that is, ka and kd, as outputs, were obtained through fitting the reconstructed ideal sensorgram. The results showed that the prediction errors of ka and kd obtained by our model were less than 12 and 24%, respectively. Based on the convenience, accuracy, and reliability of the proposed DL approach, we believe our strategy significantly boosts the feasibility to monitor the binding affinity of antibodies online during production.
近年來,人工智慧的深度學習(DL)維度受到生化研究人員的廣泛關注,逐漸成為生物感測應用領域採用的關鍵方法。研究表明,利用深度學習技術進行感測不僅可以縮短數據分析時間,還可以顯著提高數據分析和預測的準確性,從而使生物感測系統的性能較傳統方法有所提高。然而,迄今為止,在檢測過程中獲得可靠的生物分子相互作用的平衡和速率常數仍然很困難且耗時。在本研究中,我們提出了一個基於深度遷移學習和序列到序列自動編碼器的轉換模型,可以成功地將SPR感測圖轉換為蛋白質結合常數,即結合速率常數(ka)和解離速率常數(kd),它為理解藥物作用機制和生物分子的功能結構提供了重要資訊。在實驗上,我們首先使用 Langmuir 模型訓練和測試預訓練模型,該模型產生理想的 SPR 感測圖,然後透過使用合成少數過採樣技術 (SMOTE) 合成的增強 SPR 感測圖對預訓練模型進行微調透過中等規模的實驗。接下來,向微調模型輸入僅需要110 s的短實驗SPR感測圖,並將此感測圖直接轉換為重建的理想感測圖。最後,透過擬合重建的理想感測圖獲得結合動力學常數,即k a和k d作為輸出。結果表明,我們的模型獲得的k a和k d的預測誤差分別小於 12% 和 24%。基於所提出的深度學習方法的便利性、準確性和可靠性,我們相信我們的策略顯著提高了在生產過程中在線監測抗體結合親和力的可行性。